Observed size spectra
In this tutorial you will take observational data and plot the resulting size spectra for the community and for individual species. This will give you a concrete understanding of what size spectra are. There are various different ways of how size spectra can be represented, and this is a common source of confusion. We will discuss this in detail.
Introduction
We will start the introduction into size spectra using text from Ken H Andersen’s book “Fish Ecology, Evolution, and Exploitation” (2019):
What is the abundance of organisms in the ocean as a function of body size? If you take a representative sample of all life in the ocean and organize it according to the logarithm of body size, a remarkable pattern appears: the total biomass of all species is almost the same in each size group. The sample of marine life does not have to be very large for the pattern to appear. … What is even more surprising is that the pattern extends beyond the microbial community sampled by plankton nets—it persists up to the largest fish, and even to large marine mammals.
This regular pattern is often referred to as the Sheldon spectrum in deference to R. W. Sheldon, who first described it in a ground-breaking series of publications. Sheldon had gotten hold of an early Coulter counter that quickly and efficiently measured the size of microscopic particles in water. Applying the Coulter counter to microbial life in samples of coastal sea water, he observed that the biomass was roughly independent of cell size among these small organisms (Sheldon and Parsons, 1967). And he saw the pattern repeated again and again when he applied the technique to samples from around the world’s oceans. Mulling over this result for a few years, he came up with a bold conjecture (Sheldon et al., 1972): the pattern exists not only among microbial aquatic life, but it also extends all the way from bacteria to whales.
You can read more about this work in the references below:
Sheldon, R. W., and T. R. Parsons (1967). “A Continuous Size Spectrum for Particulate Matter in the Sea.” Journal Fisheris Research Board of Canada 24(5): 909–915.
Sheldon, R.W., A. Prakash, andW. H. Sutcliffe (1972). “The Size Distribution of Particles in the Ocean.” Limnology and Oceanography 17(3): 327–340
Blanchard, J. L., Heneghan, R. F., Everett, J. D., Trebilco, R., & Richardson, A. J. (2017). From bacteria to whales: using functional size spectra to model marine ecosystems. Trends in ecology & evolution, 32(3), 174-186.
K.H. Andersen, N.S. Jacobsen and K.D. Farnsworth (2016): The theoretical foundations for size spectrum models of fish communities. Canadian Journal of Fisheries and Aquatic Science 73(4): 575-588.
K.H. Andersen (2019): Fish Ecology, Evolution, and Exploitation - a New Theoretical Synthesis. Princeton University Press.
And in many other references listed in the sizespectrum.org publications page
Example code
These tutorials contain a lot of working R code, because learning R coding for mizer is best done by looking at example code. We envisage that you may want to copy and paste the code from this tutorial into your tutorial worksheet to try it out and possibly modify it to see what happens.
Now that the live version of this course has finished, you no longer need to use the worksheets mentioned below. It is not guaranteed that they will still work. You are still encouraged to try out the code that you find in the tutorials on this website and to complete the exercises but you do not need these worksheets for that purpose. All the code you need is given in the tutorials.
To analyse and plot the data we will be making use of the tidyverse package, in particular dplyr and ggplot2. If you are not familiar with these, you can learn what is needed just by studying the example code in this tutorial. Before we can use a package we need to load it in from the package library with the library() function:
When you hover over any function name in the R code in these tutorials, you will notice that they are links. If you click on them this will open the function’s help page in a new browser window. Thus you can always easily read more information about the functions we are using.
You will find explanations of the code if you expand the “R details” sections located below many of the code chunks. To expand an explanation section, just click on the “Expand for R details”.
The data
So let’s see if we can find Sheldon’s pattern ourselves. First we load the data.
download.file("https://github.com/sizespectrum/mizerCourse/raw/main/understand/size-data.rds",
destfile = "size-data.rds")'data.frame': 925594 obs. of 3 variables:
$ species: chr "Cod" "Cod" "Cod" "Cod" ...
$ weight : num 52.77 56.37 6.14 5.66 5.89 ...
$ length : num 17.41 17.8 8.5 8.27 8.38 ...The data consists of measurements of the length in centimetres of 925594 fish of various species. The species included are
unique(data$species)[1] "Cod" "Whiting" "Haddock" "Saithe" "Herring" "Sandeel"
[7] "Nor. pout" "Plaice" "Sole" This data was assembled by Ken Andersen at DTU Aqua in Copenhagen. The length in centimetres l was converted to weight in grams w using the standard allometric relationship
w = a\, l ^ b,
where the coefficient a and the exponent b are species-specific parameters (we’ll discuss how to find such species parameters in a later tutorial). The reason we like to work with weight as the measure of a fish’s size is that there are well-known allometric relationships between weight and physiological rates. For example, metabolic rate is generally expected to scale as w^{3/4} and mortality to scale as w^{-1/4}, as we will be discussing in the section on allometric rates in the next tutorial.
When not otherwise specified, all lengths are given in centimetres [cm] and all weights are given in grams [g].
Histogram
To get an impression of the size distribution of the fish, we plot a histogram of the fish weights.
p <- ggplot(data) +
geom_histogram(aes(weight), fill = "blue", colour = "black") +
scale_x_continuous(name = "Weight [g]") +
scale_y_continuous(name = "Number of fish")
p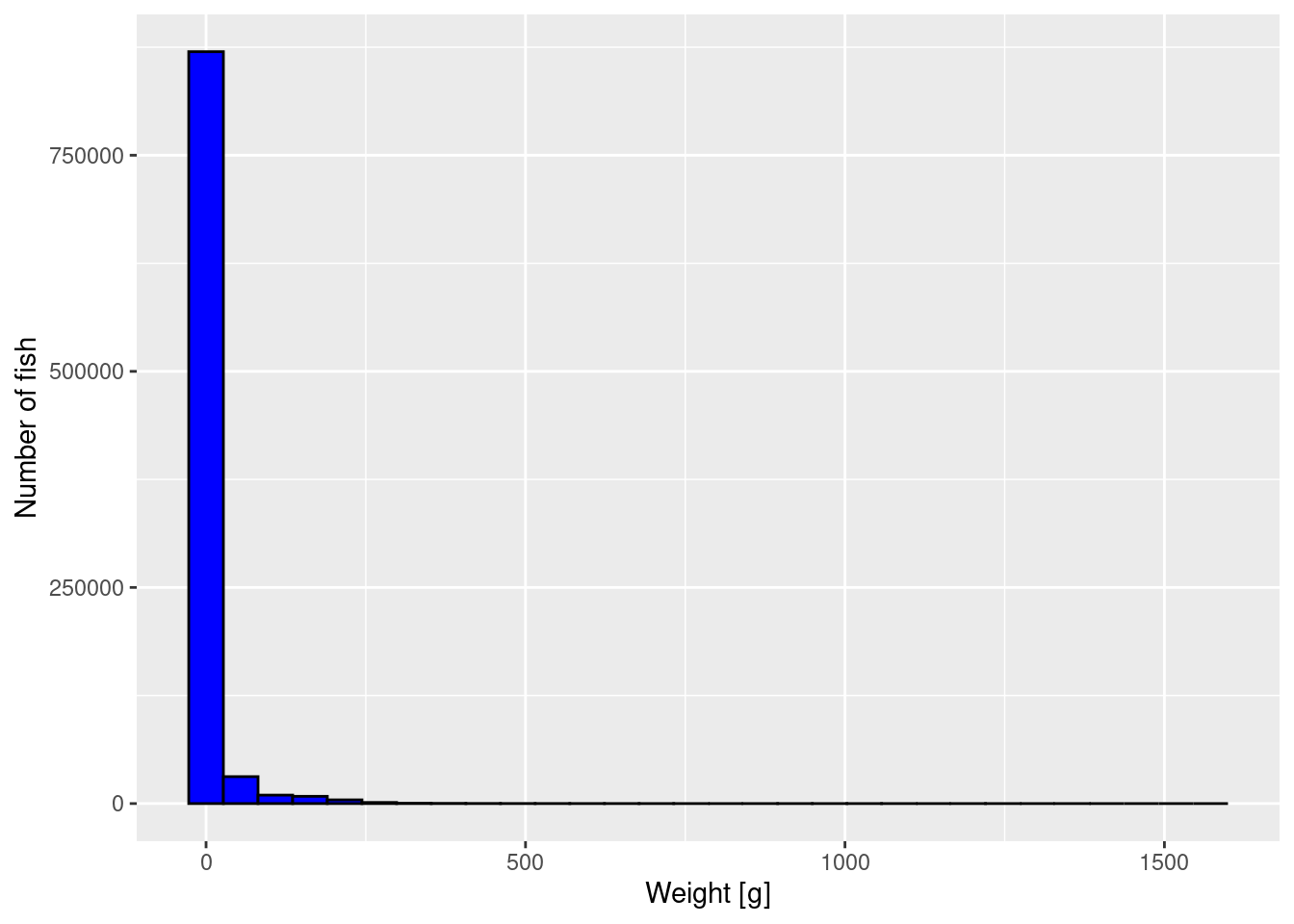
We have used the ggplot2 package, included in the tidyverse package, to make the plot. It is much more powerful and convenient than the base plot commands. It implements the “grammar of graphics”. If you are not already using ggplot2 it is worth your time to familiarise yourself with it. However for the purpose of this tutorial, you can simply pick up the syntax from the examples we give.
In the above we first specify with ggplot(data) that the graph shall be based on the data frame data that we loaded previously. We then add features to the graph with the +.
First geom_histogram() specifies that we want a histogram plot. The argument specifies the variable to be represented. Note how this is wrapped in a call to aes(). Don’t ask why, that is the way the grammar of graphics likes it. The specification of how the variables are tied to the aesthetics of the graph will always be given withing the aes() function.
We then specify that we want the bars in the histogram to be blue (fill = "blue") with a black border (colour = "black"). Such tuning of the appearance is of course totally optional. By the way: one has to admire how the ggplot2 package accepts both colour and color, so that our US friends can use color = "black".
Then we add our own labels to the axes with labs().
We assign the resulting plot to the variable p because that way we can manipulate the plot further below. Because the assignment operator in R does not display any result, we added another line with just the variable name p which will display the plot.
The plot is not very informative. It just tells us that most fish are very small but there is a small number of very large fish. We can not see much detail. We will apply 3 methods to improve the graph.
Logarithmic y axis
The first way to improve the plot is to plot the y-axis on a logarithmic scale. That has the effect of stretching out the small values and squashing the large values, revealing more detail.
p + scale_y_log10()Warning: Transformation introduced infinite values in continuous y-axisWarning: Removed 2 rows containing missing values (`geom_bar()`).
We get a warning because there were bins that contained no fish, and taking the log of 0 is not allowed. We can ignore these warnings because the empty bins will simply not be given a bar in the resulting plot.
Logarithmically sized bins
The second way to deal with the fact that there are so many more small fish than large fish, is to make the bin widths different. At the moment all bins are the same width, but we can make bin widths larger at larger sizes and smaller at smaller sizes. For example we could make the smallest bin go from 1 gram to 2 gram, the next bin to go from 2 gram to 4 gram, and so on, with each next bin twice the size of the previous. This means that for large fish bins will be very wide and a lot more individuals will fall into these bins. So let’s create the break points between these bins:
(log_breaks <- seq(from = 0, to = 11, by = 1)) [1] 0 1 2 3 4 5 6 7 8 9 10 11(binbreaks <- 2 ^ log_breaks) [1] 1 2 4 8 16 32 64 128 256 512 1024 2048We had decided that we wanted the breaks between the bins to be located at powers of 2. We first create the vector log_breaks with the exponents. The seq() function creates a vector of numbers starting at from and going up to to in steps of size by. You do not need to give the names of the arguments because they can also be identified by their position. So you could also have written seq(0, 11, 1). Such integer sequences are used so often that there is the even shorter notation 0:11 giving the same sequence.
The second line above creates a vector containing the powers of 2 with the exponents we just created in the vector log_breaks. Note how R can perform calculations for all entries of a vector at once, without the need for a loop.
The reason we have put parentheses ( ... ) around the assignments in the above code is because that also leads to the result being displayed whereas the assignment operator without those parentheses around it would not display anything.
Now we can use these logarithmically spaced bins in the histogram while also keeping the logarithmic y-axis.
p2 <- ggplot(data) +
geom_histogram(aes(weight), fill = "blue", colour = "black",
breaks = binbreaks) +
labs(x = "Weight [g]",
y = "Number of fish") +
scale_y_log10()
p2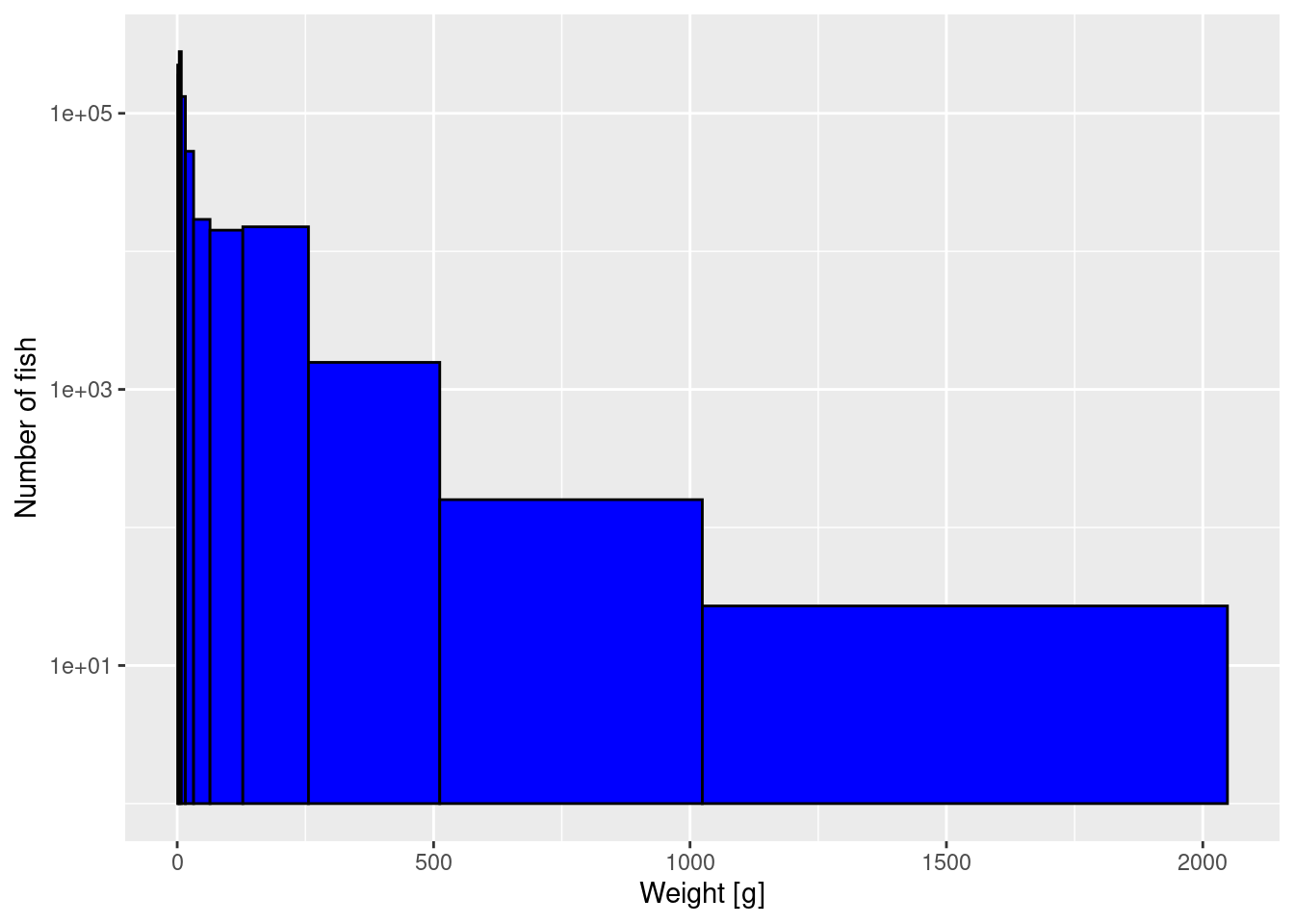
The heights are now slightly more even among the bins, because the largest bin is so wide.
It is very important that you break up your reading of the tutorials with some hands-on work that builds on what you have just learned. Therefore you will find exercises throughout the tutorials. You have now reached the first exercise. You will find that it can be accomplished using the R functions you have already learned about. Please do your work in the worksheet that accompanies this tutorial and that you will find in this week’s worksheet repository.
Now that the live version of this course has finished, you no longer need the worksheet repository mentioned below. Instead you can simply create your own worksheets by copying code over from the tutorials.
If you have not yet created this week’s worksheet repository, you can do so by following this link to accept your assignments for week 1:
https://classroom.github.com/a/5cnr1H7R
Once your repository has been created on GitHub, you will be given the URL that you need to clone it to your computer by creating a new project in RStudio. To do that you will need to follow the same steps as in the Use Git and GitHub tutorial. You may want to review the instructions in the Create repository and Clone repository sections.
You will find the worksheet for this tutorial in the file “worksheet1-observed-size-spectra.Rmd” in your worksheet repository for this week. In that worksheet you will find the following first exercise:
Now you want to double the number of logarithmically-sized bins used to get a more detailed picture of the size spectrum. So instead of using 11 bins to cover the range from 1g to 2048g, where each bin is twice as large as the previous, you want to use 22 bins where each bin is larger than the previous one by a factor of \sqrt{2}.
Create the vector with the breaks between the bins and then use that when you plot the histogram.
After successful completion of that exercise, return here to continue with the tutorial. Do not do the other exercises until you reach them in this tutorial.
Logarithmic w axis
Finally, we can also display the weight axis on a logarithmic scale:
p2 + scale_x_log10()
Note how on the logarithmic axis the logarithmically-sized bins all have the same width.
Densities
Note that the height of the bars changed as we changed how we bin the data. That is obvious. If we make a bin twice as large, we expect twice as many fish in that bin.
Now please pay attention! We want to get rid of this dependence on the choice of the bin width. We want to represent the data in a way that does not depend on our choice of binning. We can do this if we divide the height of each bar by its width. This means we will work with density and the bin height will be scaled by the bin width.
Because it is important to understand this concept of density, we will calculate the number density by hand below, even though ggplot has a built-in function geom_density() that we could use instead. We first bin the data by hand, and then we calculate and plot the densities.
Binning
To understand better what the histogram did and to improve the plots further, we bin the data ourselves. We do this by first adding a bin number to each observation, which indicates in which bin the weight of the fish lies.
data_with_bins <- data |>
mutate(bin = cut(weight, breaks = binbreaks, right = FALSE,
labels = FALSE))
head(data_with_bins)We used the pipe operator |> that simply pipes the output of the code preceeding it into the first argument of the function following it. So the above code is equivalent to
The pipe operator becomes really useful only if you do a longer sequence of operations on data. You will see examples of its use later.
The mutate() function can add new columns to a data frame or modify existing columns. In the above example it adds a new column bin. The entries in that column are here calculated by the function cut that returns the label of the bin into which an observation falls. We specify the bin boundaries with the breaks = binbreaks to be the boundaries we have calculated above. The right = FALSE means that in case an observation falls exactly on a right bin boundary, it is not included in that bin but instead in the next bin. The labels = FALSE means that the bins are not labelled by the intervals but simply by integer codes.
We then group the data by bin and calculate the number of fish in each bin
After we have grouped together all the observations with the same bin number with the group_by(bin), the summarize() function creates a new data frame with one row for each group, which in this case means one row for each bin. That data frame will always have one column specifying the group and then we specified that we want an extra column Numbers that just counts the number of observations in the group with the n() function. Note that the species is ignored in this calculation.
In the above code you see the pipe operator |> being quite convenient, because it allows us to write the functions in the order in which they are applied, rather than having to write summarize(group_by(...)).
The numbers in each bin give us the heights of the histogram above.
Number density
The values for Numbers of course depend on the size of the bins we have chosen. Wider bins will have more fish in them. It is therefore convenient to divide these numbers by the bin widths to get the average number density in the bins.
We first calculate the widths of the bins using the diff() function which calculates the difference between neighbouring entries in a vector. Then when we calculate the entries for the new Number_dens column we pick out the bin width appropriate to the given bin for each observation.
Let’s make a plot of the number density against weight. Note that we only estimated the number density of weights within each bin, so we only have an average value which we use to determine the height of the curve at the midpoint of the bin. So the plot interpolates between these discrete points (at the midpoint of each bin weights) by straight lines to produce a continuous curve.
bin_midpoints <- 2 ^ (log_breaks[-length(log_breaks)] + 1/2)
binned_numbers$bin_midpoint = bin_midpoints
ggplot(binned_numbers) +
geom_line(aes(x = bin_midpoint, y = Number_dens)) +
scale_x_continuous(name = "Weight [g]") +
scale_y_continuous(name = "Number density")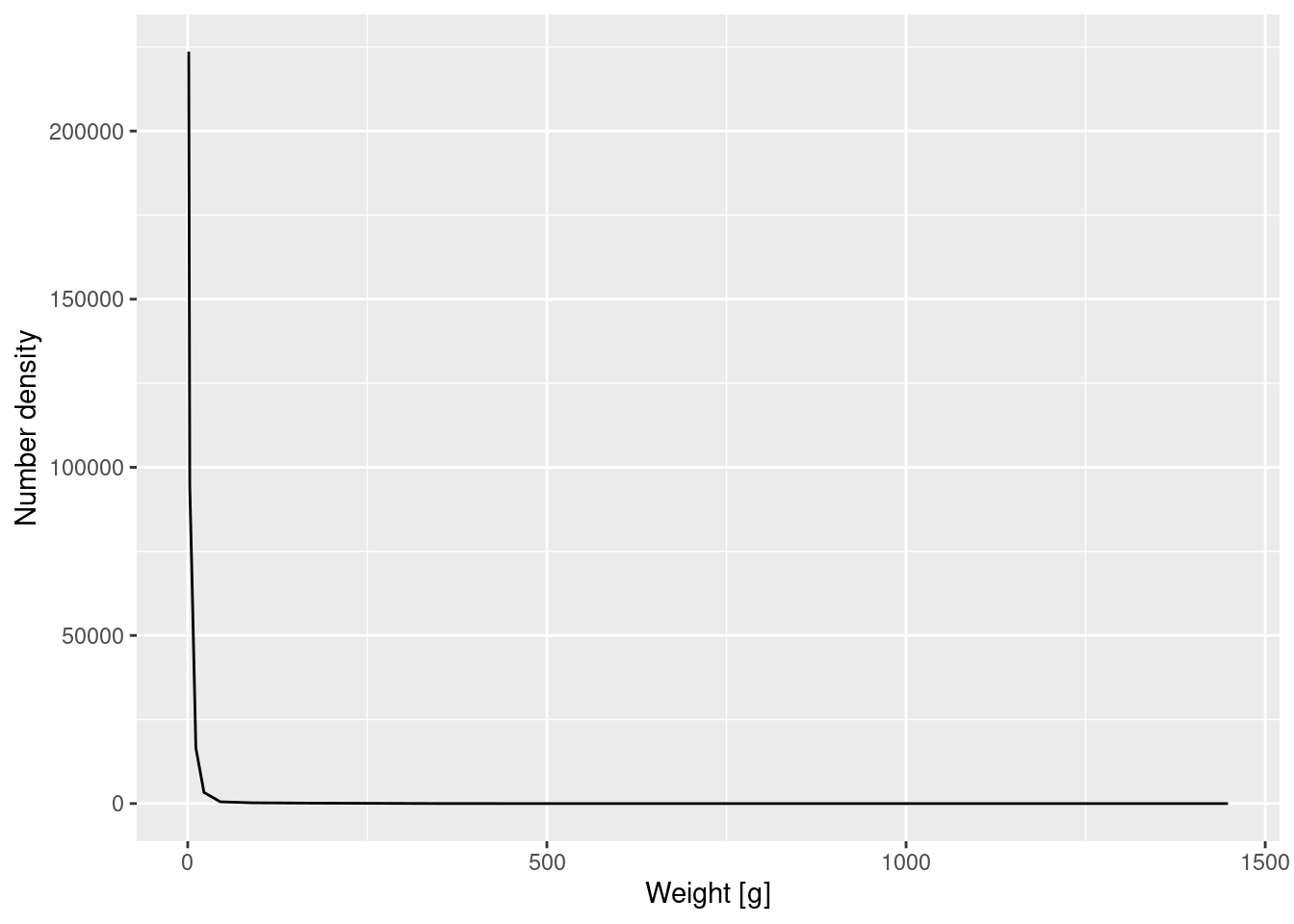
We first calculate a vector bin_midpoints holding the midpoint of all the bins. We do this using the vector log_breaks[-length(log_breaks)] which contains all the entries of log_breaks except the last. We then add a bin_midpoint column to the dataframe binned_numbers and use that as the variable for the x-axis in the plot.
Again the graph tells us that most of the individuals are very small, but we can not see any of the details. We therefore plot the density on log-log axes:
ggplot(binned_numbers) +
geom_line(aes(x = bin_midpoint, y = Number_dens)) +
scale_x_log10(name = "Weight [g]") +
scale_y_log10(name = "Number density")
It is time for your second exercise of the course. Go back to your week 1 exercise project in RStudio, and do Exercise 2 in worksheet 1:
Earlier you increased the number of bins from 11 to 22. Because the same number of observed fish was then spread over this larger number of bins, all the bars in the histogram were accordingly less high. By going to the number density we have corrected for this. The density plot created with the 22 bins will of course not look exactly the same as the one created with 11 bins. It will look more ragged because it exposes the noise in the data more.
Create the plot of the number density using the 22 logarithmically sized bins from exercise 1.
Fitting a power law
The number density in the above log-log plot is described approximately by a straight line. We can approximate the slope and intercept of that straight line by fitting a linear model
Call:
lm(formula = log(Number_dens) ~ log(bin_midpoint), data = binned_numbers)
Coefficients:
(Intercept) log(bin_midpoint)
14.563 -2.241 This tells us that the straight-line approximation has a slope of about -2.24. We can also ask ggplot to put this line into the plot, together with its 95% confidence interval:
ggplot(binned_numbers, aes(x = bin_midpoint, y = Number_dens)) +
geom_line() +
scale_x_log10(name = "Weight [g]") +
scale_y_log10(name = "Number density") +
geom_smooth(method = 'lm')`geom_smooth()` using formula = 'y ~ x'
The linear regression line is produced by geom_smooth(method='lm'). Note how we moved the call to aes() into the call to ggplot(). That is then used automatically for both the geom_line() and the geom_smooth(), so that we did not have to specify the information twice.
If we denote the number density at weight w by N(w), then the above tells us that \log(N(w)) \approx 14.6 + \log(w)^{-2.24}.
If we exponentiate both sides to get rid of the logarithms this gives
N(w) \approx \exp(14.6) w^{-2.24} = N(1) w^{-\lambda}
with \lambda \approx 2.24.
A straight line on a log-log plot indicates a power-law relationship between the variables with the slope of the line being the exponent in the power law.
The term lambda or \lambda is widely used in size spectrum terminology and it denotes the size spectrum slope. A steeper slope (larger \lambda value) means that there are relatively fewer large fish compared to small fish. A more shallow slope (smaller \lambda) indicates a relatively larger number of large fish. Now you know how these slopes are calculated.
Of course the approach we took above of estimating the exponent in the power law from the binned data is not ideal. If one has access to unbinned data, as we have here, one should always use that unbinned data. So the better way to estimate the slope or exponent from our data would be to ask: “If we view our set of observed fish sizes as a random sample from the population described by the power law, for which exponent would our observations be the most likely”. In other words, we should do a maximum likelihood estimation of the exponent. We’ll skip the mathematical details and just tell you the result that the maximum likelihood estimate for the power-law exponent \lambda is \lambda = 1+\frac{n}{\sum_{i=1}^n\log\frac{w_i}{w_{min}}}, where n is the number of observed fish, w_i is the weight of the ith fish and w_{min} is the weight of the smallest fish. For our data this gives
[1] 1.709584You can see that this approach, which gives equal weight to each observation rather than giving equal weight to each bin, gives a lower value for \lambda, namely \lambda \approx 1.71 instead of \lambda \approx 2.24. This is quite a big difference. But this is still not the end of the story, because we did not take measurement error into account. We assumed that we sampled perfectly. But in reality, small individuals are much easier to miss than large ones, so our data is almost certainly under-reporting the number of small individuals, which leads to a smaller \lambda or more shallow size spectra. Also, in many ecosystems large fish have been removed by fishing, so we might also be missing them. This would lead to steeper slopes and larger \lambda. But this is a separate set of analsyes.
To estimate slopes properly we should try to observe the power law over a wider range of sizes, all the way from bacteria to whales. This is what Sheldon et.al. did in 1972 and he observed that \lambda \approx 2. More thorough investigations since then have led to values just slightly above 2 on average. TODO: references.
Biomass density
Above we first calculated the number of fish in each weight bin and then divided by the width of each bin to obtain the average number density in each bin. Exactly analogous to that, we can calculate the biomass of all the fish in each weight bin and divide that by the width of each bin to obtain the average biomass density in each bin. Note, that number density and biomass density will give quite different results, because there are lots of fish in small bins but their total biomass may not be very large. So in the code below we will now sum weight and not numbers.
Make a plot of the biomass density similarly to how we plotted the number density above, using logarithmic axes and also plotting the straight-line approximation.
Fitting a same linear model to the binned biomass density data now gives
Call:
lm(formula = log(Biomass_dens) ~ log(bin_midpoint), data = binned_biomass)
Coefficients:
(Intercept) log(bin_midpoint)
14.615 -1.265 The slope is -1.265 for the biomass density whereas it was -2.24 for the number density. This makes perfect sense, because if we denote the biomass density by B(w) then B(w) = w N(w). Therefore if the number density scales with the exponent \lambda as N(w) \propto w^{-\lambda} then the biomass density will scale as B(w)\propto w^{1-\lambda}.
Densities in log weight
So far we found various size spectrum slopes and evidence of decreasing biomass densities with size. Yet, the tutorial started with a reference to Sheldon’s observation about equal biomass across sizes. Why?
Above, we calculated densities by dividing the total number or total biomass in each bin by the width that the bin has on the linear weight (w) axis. Instead, we could divide the total biomass in each bin by the width of the bin on the logarithmic weight axis.
We saw earlier that when we plot the logarithmically-sized bins on a logarithmic w axis they are all equally sized. We can calculate the size of the bins in log weight:
[1] 0.30103 0.30103 0.30103 0.30103 0.30103 0.30103 0.30103 0.30103 0.30103
[10] 0.30103 0.30103We used the logarithm to base 10 above because that is the convention that mizer uses.
An aside: Because, while we wrote this section we got confused ourselves, let us state an obvious fact lest it cause confusion for you as well: the width of a bin on the logarithmic axis is not the logarithm of the width of the bin on the linear axis.
Now if we divide the number of fish in each bin by the width of the size bin in log weight we get what we denote as the “number density in log weight”. Similarly if we divide the biomass in each bin by the width of the size bin on a logarithmic axis we get the “biomass density in log weight”, which is the Sheldon density.
Below we plot the number densities (in black) and the biomass densities (in blue), with the solid lines the densities in weight and the dashed lines the densities in log weight, all on the same axes.
# remember how we calculated densities by dividing numbers and biomasses
# by bin_widths
binned <- data_with_bins |>
group_by(bin) |>
summarise(Numbers = n(),
Biomass = sum(weight)) |>
mutate(bin_midpoint = bin_midpoints,
Number_dens = Numbers / bin_width[bin],
Biomass_dens = Biomass / bin_width[bin])
# and now we add densities in log weight by dividing by log_bin_width
binned <- binned |>
mutate(Number_dens_log_w = Numbers / log10_bin_width[bin],
Biomass_dens_log_w = Biomass / log10_bin_width[bin])
ggplot(binned) +
geom_line(aes(x = bin_midpoint, y = Number_dens),
colour = "black") +
geom_line(aes(x = bin_midpoint, y = Biomass_dens),
colour = "blue") +
geom_line(aes(x = bin_midpoint, y = Number_dens_log_w),
colour = "black", linetype = "dashed") +
geom_line(aes(x = bin_midpoint, y = Biomass_dens_log_w),
colour = "blue", linetype = "dashed") +
scale_x_log10(name = "Weight [g]") +
scale_y_log10(name = "Density")
The blue dashed line shows that Sheldon’s density. You will notice that the slope of the Sheldon density is less negative than the slope of the biomass density.
We introduce the notation N_{\log w}(w) for the number density in log weight and B_{\log w}(w) for the biomass density in log weight (Sheldon’s density). We have the following relations among the various densities:
B_{\log w}(w) = w\, B(w) \propto w\, N_{\log}(w) = w^2 N(w).
Sheldon’s observation was that B_{\log w}(w) is approximately constant over a large range of sizes w from bacteria to whales. That corresponds to the earlier phrasing of Sheldon’s observation that N(w)\propto w^{-\lambda} with \lambda close to 2. Here is a very important and interesting summary about the slopes:
The slope of the number density is approximately -2, the slope of the biomass density and the number density in log weight is approximately -1, and the slope of the biomass density in log weight is approximately 0, i.e., the biomass density in log weight is approximately constant.
When we talk of “log weight”, we have to define what we mean. Weight is a dimensionful quantity. We need to choose a unit before we can take the logarithm. In mizer we choose to measure weight in grams. So when we say “log weight”, we mean the logarithm of the weight measured in grams. The result is a dimensionless quantity.
We also need to specify the base of the logarithm. The ggplot package likes to work with log10, mizer also follows that convention, therefore that is what we chose in the above graphs. It does not matter: choosing a different base just rescales the density by an overall factor.
The different densities have different units. This is the result of two factors: 1) Biomass is measured in grams whereas Numbers are dimensionless and 2) bin widths are measured in grams whereas bin widths in log weight are dimensionless. The densities, obtained by dividing either Biomass or Numbers by either bin widths or bin widths in log weight, have the following units:
- N(w) has unit 1/grams,
- B(w) and N_{\log w}(w) are dimensionless,
- B_{\log w}(w) has unit grams.
We know that one should be careful when plotting quantities with different dimensions on the same axis because the result depends on the units chosen. Above we chose grams as our unit.
Size spectra of individual species
So far we have looked at the community spectrum, where we ignored the species identity of the fish. We will now look at the spectra of the individual species. We’ll plot them all on the same graph and display them with plotly so that you can hover over the resulting graph to see which line corresponds to which species. We also include the community size spectrum in black for comparison. The lines look smoother than earlier because now we use kernel density estimation rather than binning to estimate the densities.
p <- ggplot(data) +
geom_density(aes(weight, stat(count), colour = species), adjust = 4) +
geom_density(aes(weight, stat(count)), colour = "black", lwd = 1.2, adjust = 4) +
scale_x_continuous(trans = "log10", name = "Weight [g]") +
scale_y_continuous(trans = "log10", limits = c(1, NA), name = "Number density in log w")
plotly::ggplotly(p)Warning: `stat(count)` was deprecated in ggplot2 3.4.0.
ℹ Please use `after_stat(count)` instead.
ℹ The deprecated feature was likely used in the ggplot2 package.
Please report the issue at <https://github.com/tidyverse/ggplot2/issues>.Warning: Transformation introduced infinite values in continuous y-axisBinning the data is useful but is not the only way to approximate the densities. In reality you don’t need to bin the data by hand or bin it at all. One can also use the kernel density estimation method and ggplot2 even has that built in to its geom_density().
By default, geom_density() would normalise the density so that the integral under the density curve is 1. We use stat(count) to tell it that we want the number density, so that the integral under the curve is the total number, not normalised to 1.
The kernel density method works by placing a small Gaussian (bell curve) at each data point and then adding up all these little Gaussians. The adjust = 4 means that the width of these Gaussians is 4 times wider than geom_density() would choose by default. This leads to a smoother density estimate curve. This plays a similar role as the bin width does in histograms.
Note how easy it was to ask ggplot to draw a separate line for each species. We only had to add the colour = species to geom_density(). ggplot automatically chose some colours for the species and added a legend to the plot.
We have replaced scale_x_log10() by scale_x_continuous(trans = "log10") which does exactly the same thing. But the latter form is more flexible, which we used in scale_y_continuous(trans = "log10", limits = c(1, NA)) to also specify limits for the y axis. We chose to plot densities above 1 only because at smaller values there is only noise. We did not want to set an upper limit, hence the NA.
We then did not display the plot p but instead fed it to plotly::ggplotly(). The ggplotly() function takes a plot created with ggplot and converts it into a plot created with plotly. plotly plots are more interactive. We called the ggplotly() function with plotly::ggplotly() because that way we did not need to load the plotly package first, i.e., we did not need to do library(plotly).
There are three messages you should take away from this plot:
- Different species have very different size spectra.
- The estimates of the species size spectra are not very reliable because we do not have very good data.
- The community size spectrum looks more regular than the species size spectra.
We will discuss species size spectra more in the next tutorial, where we will look at them with the help of the mizer model.
Summary and recap
1) It is very useful to know how many organisms of different sizes there are. This is what size spectra show.
2) We can represent size spectra in different ways. One is to bin the data and plot histograms. The drawback is that the height of the bars in a histogram depend on our choice of bins. A bin from 1 to 2g will have fewer individuals than a bin from 1 to 10g.
3) To avoid the dependence on bin sizes, we use densities, where the total number of individuals or the total biomass of individuals in each bin are divided by the width of the bin. We refer to these as the number density or the biomass density respectively.
3) The number density looks very different from the biomass density. There will be a lot of very small individuals, so the number density at small sizes will be large, but each individual weighs little, so their total biomass will not be large.
5) When we work with a logarithmic weight axis, then it is natural to use densities in log weight, where the numbers of individuals in each bin are divided by the width of the bin on the log axis. We refer to these as the number density in log weight or the biomass density in log weight respectively.
6) Community size spectra are approximately power laws. When displayed on log-log axes, they look like straight lines. The slope of the number density is approximately -2, the slope of the biomass density and the number density in log weight is approximately -1, and the slope of the biomass density in log weight is approximately 0, i.e., the biomass density in log weight is approximately constant. The latter is called the Sheldon spectrum.
7) The individual species size spectra are not approximately power laws and thus do not look like straight lines on log-log axes. The approximate power law only emerges when we add all the species spectra together.